




Compute degree days in R
https://ucanr-igis.github.io/degday/
Data management utilities for drone mapping
https://ucanr-igis.github.io/uasimg/

Bring climate data from Cal-Adapt into R using the API
https://ucanr-igis.github.io/caladaptr/
Homerange and spatial-temporal pattern analysis for wildlife tracking
data
http://tlocoh.r-forge.r-project.org/
Chill Portions Under Climate Change Calculator
https://ucanr-igis.shinyapps.io/chill/
Drone Mission Planner for Reforestation Monitoring Protocol
https://ucanr-igis.shinyapps.io/uav_stocking_survey/
Stock Pond Volume Calculator
https://ucanr-igis.shinyapps.io/PondCalc/
Pistachio Nut Growth Calculator
https://ucanr-igis.shinyapps.io/pist_gdd/

Whatever is needed to get your data frame ready
for the function(s) you want to use for analysis and visualization.
Also called: data munging, data manipulation, data transformation, etc.

Data wrangling often includes some combination of:
Developing a strategy
R Methods
More advanced tasks
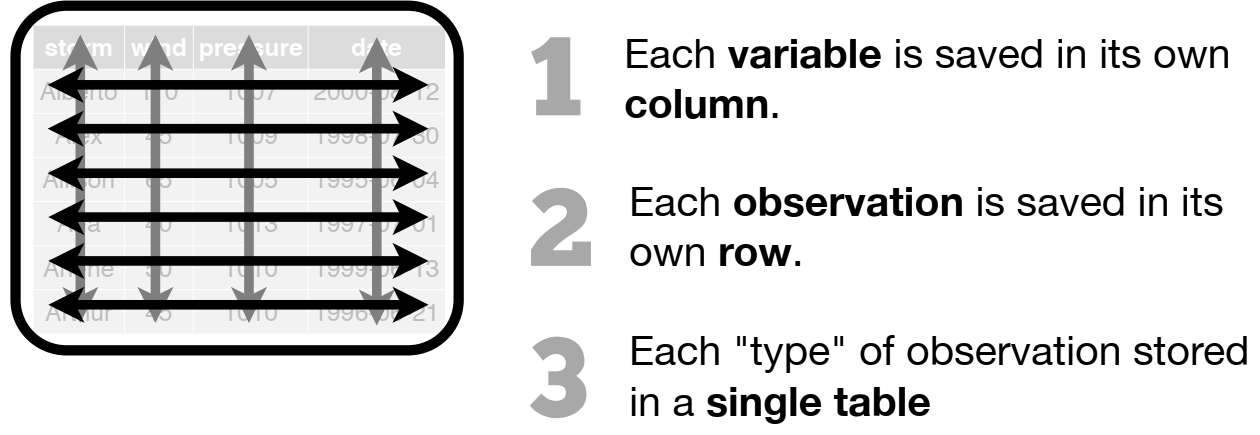
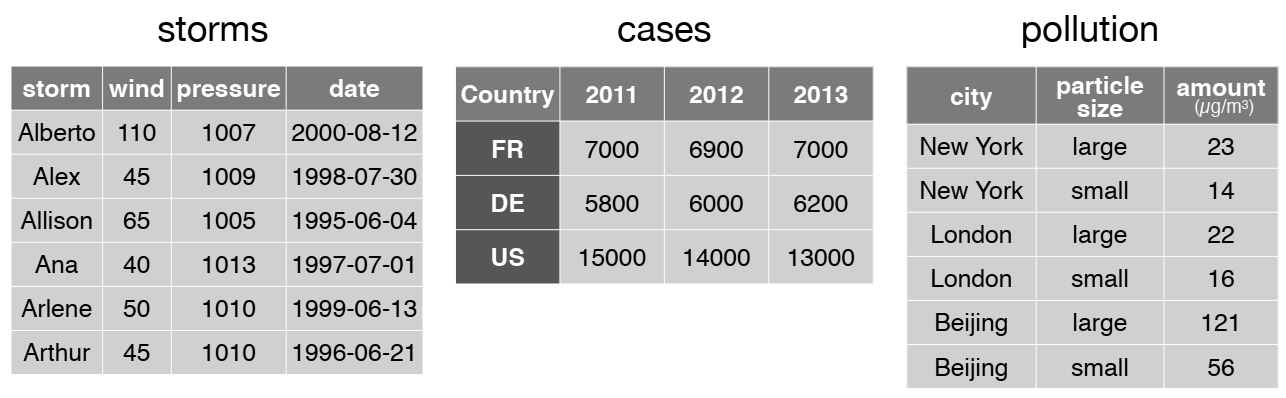
Wickham, H. (2014). Tidy data. Journal of statistical software, 59, 1-23. https://doi.org/10.18637/jss.v059.i10

Install them all at once:
For the complete list of all the tidyverse packages, see:




These packages allow you to:
Example:
So whatdya we got here?
dim()
nrow()
ncol()
str()head()
View()
dplyr::glimpse()summary()
## Discrete values
unique()
table()hist()
plot(x,y)
qqplot()is.na()
anyNA()summary()dplyrAn alternative (usually better) way to wrangle data frames than base R.
Best way to familiarize yourself - explore the cheat sheet:
dplyr Functions| subset rows | filter(), slice() |
| order rows | arrange() |
| pick column(s) | select(), pull() |
| add new columns | mutate() |
The two functions most commonly used to subset rows include
filter() and slice().
filter() to subset based on a logical
expressionsslice() to subset based on
indices
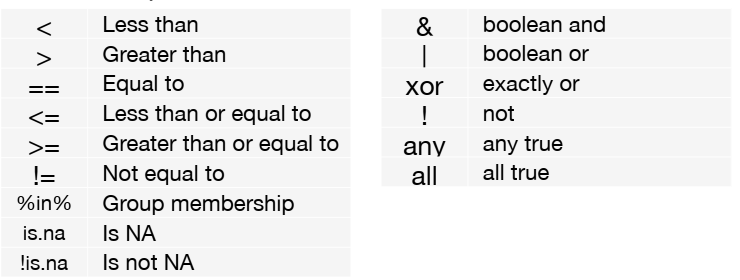
Logical expressions you can use in filter() include:
Most dplyr functions take a tibble as the first
argument , and return a tibble.
This makes them very pipe friendly.
Look at the storms tibble:
## # A tibble: 6 × 13
## name year month day hour lat long status category wind pressure
## <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <fct> <dbl> <int> <int>
## 1 Amy 1975 6 27 0 27.5 -79 tropical de… NA 25 1013
## 2 Amy 1975 6 27 6 28.5 -79 tropical de… NA 25 1013
## 3 Amy 1975 6 27 12 29.5 -79 tropical de… NA 25 1013
## 4 Amy 1975 6 27 18 30.5 -79 tropical de… NA 25 1013
## 5 Amy 1975 6 28 0 31.5 -78.8 tropical de… NA 25 1012
## 6 Amy 1975 6 28 6 32.4 -78.7 tropical de… NA 25 1012
## # ℹ 2 more variables: tropicalstorm_force_diameter <int>,
## # hurricane_force_diameter <int>
Filter out only the records for category 3 or higher storms
storms |>
select(name, year, month, category) |> ## select the columns we need
filter(category >= 3) ## select just the rows we want## # A tibble: 1,262 × 4
## name year month category
## <chr> <dbl> <dbl> <dbl>
## 1 Caroline 1975 8 3
## 2 Caroline 1975 8 3
## 3 Eloise 1975 9 3
## 4 Eloise 1975 9 3
## 5 Gladys 1975 10 3
## 6 Gladys 1975 10 3
## 7 Gladys 1975 10 4
## 8 Gladys 1975 10 4
## 9 Gladys 1975 10 3
## 10 Belle 1976 8 3
## # ℹ 1,252 more rowsdplyr functions generally allow you to enter column
names without quotes.
stringrReplace characters:
## [1] "The_Quick_Brown_Fox"
Split a character into two:
Trim white space:
ggplot2 is an extremely popular plotting package for R.
ggplots are constructed using the ‘grammar of graphics’ paradigm.







aes()
ggplot(penguins, aes(x = flipper_length_mm , y = bill_length_mm , color = species)) +
geom_point() +
ggtitle("Bill Length vs Flipper Length for 3 Species of Penguins")
aes() sets the default source for each visual property
(or aesthetic) of the plot layers
x - where it falls along the x-axis
y - where it falls along the y-axis
color
fill
size
linewidth
geom_xxxx() functions you useaes() the visual properties you want linked
to the datageom_xxxx() functions draw layers on the plot
canvas
drawn from the bottom up
some common geoms:
geom_point(col = pop_size)
geom_point(col = "red")
visual properties are inherited (from
aes())
each geom has default color palettes and legend settings
In the example below, note where geom_boxplot() gets its
visual properties:
aes()ggplot(penguins, aes(x = species, y = bill_len)) +
geom_boxplot(color = "navy", fill = "yellow", size = 1.5)## Warning: Removed 2 rows containing non-finite outside the scale range
## (`stat_boxplot()`).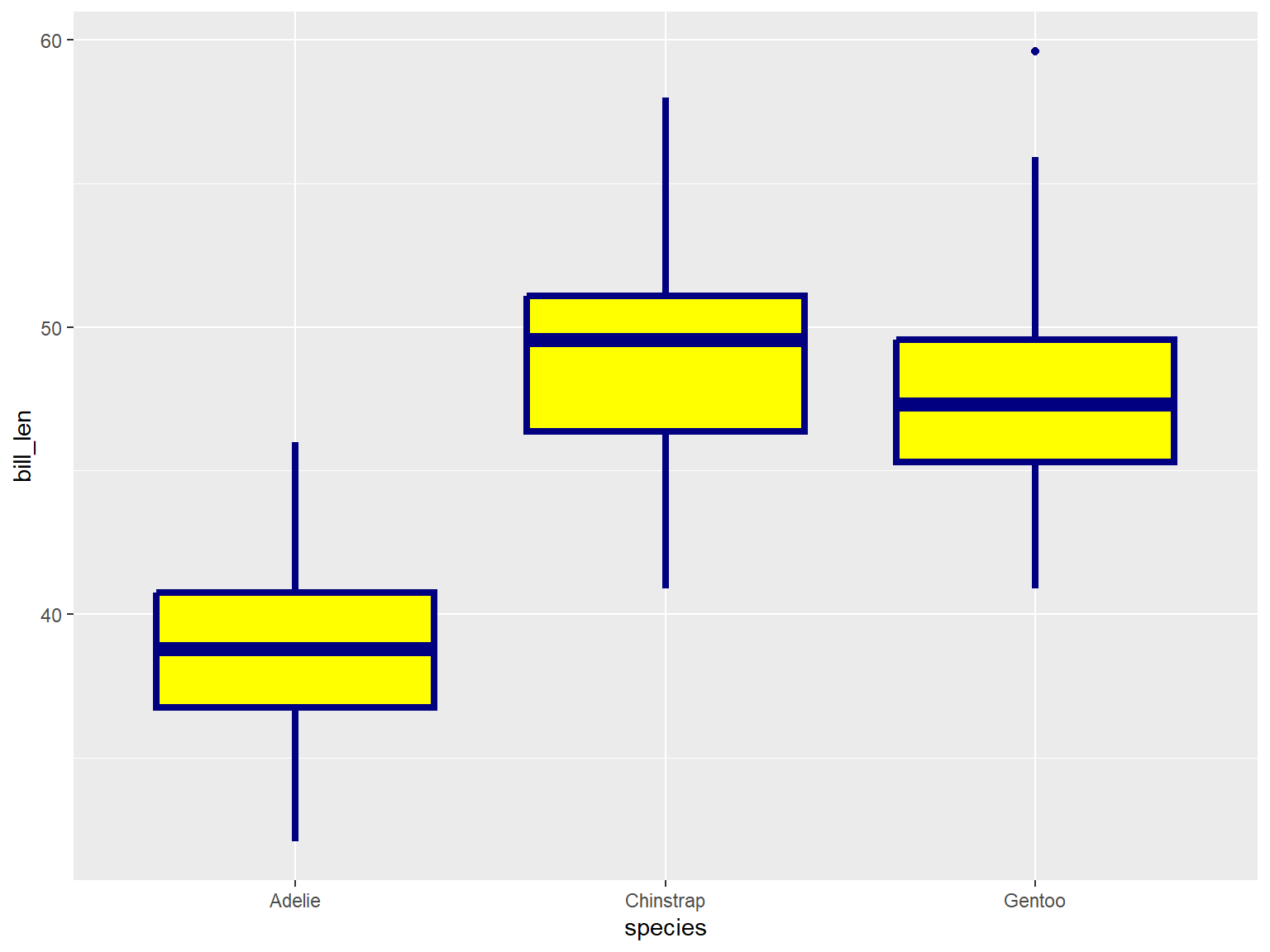
geom_xxxx() functions can also be used to add other
graphic elements:
ggplot(penguins, aes(x = species, y = bill_len)) +
geom_boxplot(color = "navy", fill = "yellow", size = 1.5) +
geom_hline(yintercept = 43.9, linewidth = 3, color="red") +
geom_label(x = 3, y = 58, label = "Gentoo penguins \n are just the best!")## Warning: Removed 2 rows containing non-finite outside the scale range
## (`stat_boxplot()`).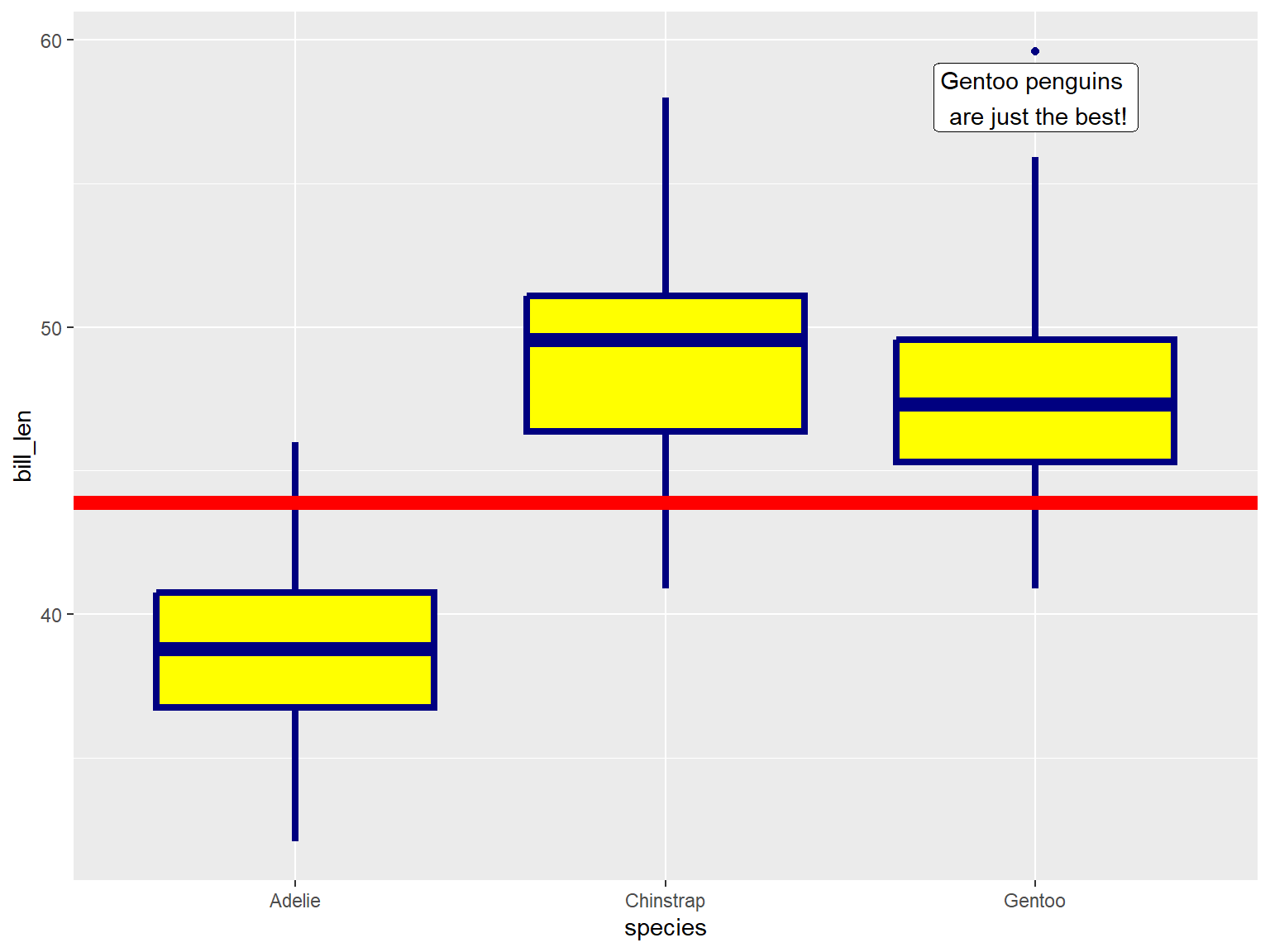
In this exercise, we will:

This exercise will be done in an Quarto Notebook!
Data wrangling starts when you import data
readxl or readrExploring what you got
ggplot() can split up the data for visualizationdim(), summarize(),
table()head(), View()Renaming columns
rename() or select()rename_with()Subsetting rows and columns
select()filter(), slice()Exploring the top and bottom values
arrange()slice_max(), slice_min()Add or modify columns: mutate()
with:
min_rank()if_else()case_when()Splitting columns
mutate + stringr::str_split_i()tidyr::separate_wider_delim() - more flexibletidy-select functions can be used to specify columns in a data frame.
These expressions can be used wherever you need to specify columns, including:
select()
rename_with()
mutate(across())
Examples
| -student_id | all columns except age |
| quiz_01:quiz10 |
all columns between quiz_01 and quiz_10
|
| starts_with("Quiz") | all columns that start with 'Quiz' |
| contains('cm') | column names that contain 'cm' |
|
all_of(c('major', 'gpa')) any_of(c('major', 'gpa')) |
column names stored in a vector |
| last_col() | last column |
| everything() | all columns |
More info:
https://tidyselect.r-lib.org/
There are better options for storing data than csv.
|
Step 1 (optional): group rows (i.e., change the unit of analysis) |
group_by() |
|
Step 2: Compute summaries for each group of rows |
Add summary columns with summarize() Aggregate functions available: n(), mean(), median(), sum(), sd(), IQR(), first(), etc. |
Example:
## # A tibble: 6 × 13
## name year month day hour lat long status category wind pressure
## <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <fct> <dbl> <int> <int>
## 1 Amy 1975 6 27 0 27.5 -79 tropical de… NA 25 1013
## 2 Amy 1975 6 27 6 28.5 -79 tropical de… NA 25 1013
## 3 Amy 1975 6 27 12 29.5 -79 tropical de… NA 25 1013
## 4 Amy 1975 6 27 18 30.5 -79 tropical de… NA 25 1013
## 5 Amy 1975 6 28 0 31.5 -78.8 tropical de… NA 25 1012
## 6 Amy 1975 6 28 6 32.4 -78.7 tropical de… NA 25 1012
## # ℹ 2 more variables: tropicalstorm_force_diameter <int>,
## # hurricane_force_diameter <int>
For each month, how many storm observations are saved in the data frame
storms |>
select(name, year, month, category) |> ## select the columns we need
group_by(month) |> ## group the rows by month
summarize(num_storms = n()) ## for each group, report the count## # A tibble: 10 × 2
## month num_storms
## <dbl> <int>
## 1 1 70
## 2 4 66
## 3 5 201
## 4 6 809
## 5 7 1651
## 6 8 4442
## 7 9 7778
## 8 10 3138
## 9 11 1170
## 10 12 212
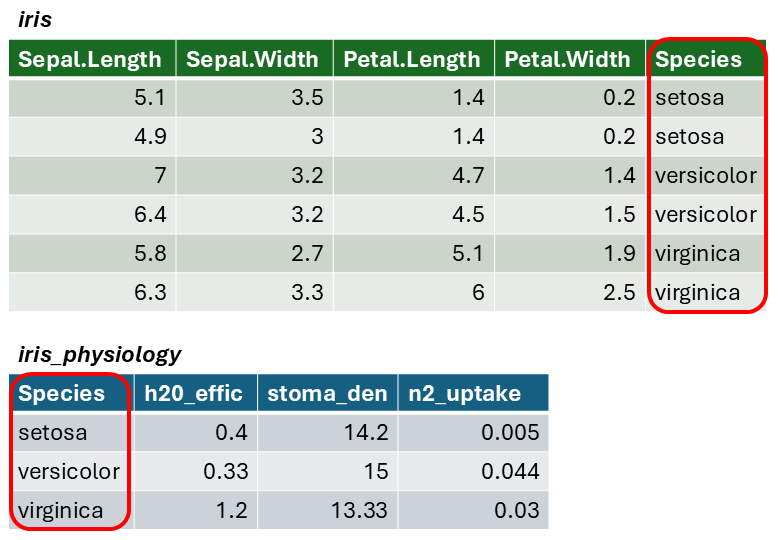
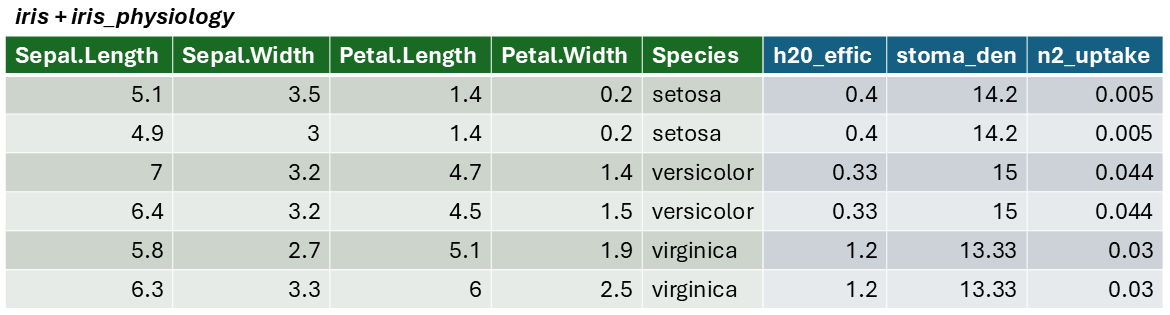
The result of a join is additional columns.
To join two data frames based on a common column, you can use:
left_join(x, y, by)
where x and y are data frames, and by is the name of a column they have in common.
Join columns must be the same data type (i.e., both numeric or both character).
If there is only one column in common, and if it has the same name in both data frames, you can omit the by argument.
The by argument can take the join_by() for more
complex cases (e.g., join columns have different names)
To illustrate a table join, we'll first import a csv with some fake data about the genetics of different iris species:
# Create a data frame with additional info about the three IRIS species
iris_genetics <- data.frame(Species=c("setosa", "versicolor", "virginica"),
num_genes = c(42000, 41000, 43000),
prp_alles_recessive = c(0.8, 0.76, 0.65))
iris_genetics## Species num_genes prp_alles_recessive
## 1 setosa 42000 0.80
## 2 versicolor 41000 0.76
## 3 virginica 43000 0.65We can join these additional columns to the iris data frame with
left_join():
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species num_genes
## 1 5.1 3.5 1.4 0.2 setosa 42000
## 2 4.9 3.0 1.4 0.2 setosa 42000
## 3 4.7 3.2 1.3 0.2 setosa 42000
## 4 4.6 3.1 1.5 0.2 setosa 42000
## 5 5.0 3.6 1.4 0.2 setosa 42000
## 6 5.4 3.9 1.7 0.4 setosa 42000
## 7 4.6 3.4 1.4 0.3 setosa 42000
## 8 5.0 3.4 1.5 0.2 setosa 42000
## 9 4.4 2.9 1.4 0.2 setosa 42000
## 10 4.9 3.1 1.5 0.1 setosa 42000
## prp_alles_recessive
## 1 0.8
## 2 0.8
## 3 0.8
## 4 0.8
## 5 0.8
## 6 0.8
## 7 0.8
## 8 0.8
## 9 0.8
## 10 0.8
In this exercise, we will continue to work with pop-quiz data, and: