



Compute degree days in R
https://ucanr-igis.github.io/degday/
Data management utilities for drone mapping
https://ucanr-igis.github.io/uasimg/

Bring climate data from Cal-Adapt into R using the API
https://ucanr-igis.github.io/caladaptr/
Homerange and spatial-temporal pattern analysis for wildlife tracking
data
http://tlocoh.r-forge.r-project.org/
Chill Portions Under Climate Change Calculator
https://ucanr-igis.shinyapps.io/chill/
Drone Mission Planner for Reforestation Monitoring Protocol
https://ucanr-igis.shinyapps.io/uav_stocking_survey/
Stock Pond Volume Calculator
https://ucanr-igis.shinyapps.io/PondCalc/
Pistachio Nut Growth Calculator
https://ucanr-igis.shinyapps.io/pist_gdd/
Whatever is needed to get your data frame ready
for the function(s) you want to use for analysis and visualization.
Also called: data munging, data manipulation, data transformation, etc.
Data wrangling often includes some combination of:

Developing a strategy
R Methods
More advanced tasks

Highlights
Suggestions for Using ChatGPT to Learn Data Wrangling
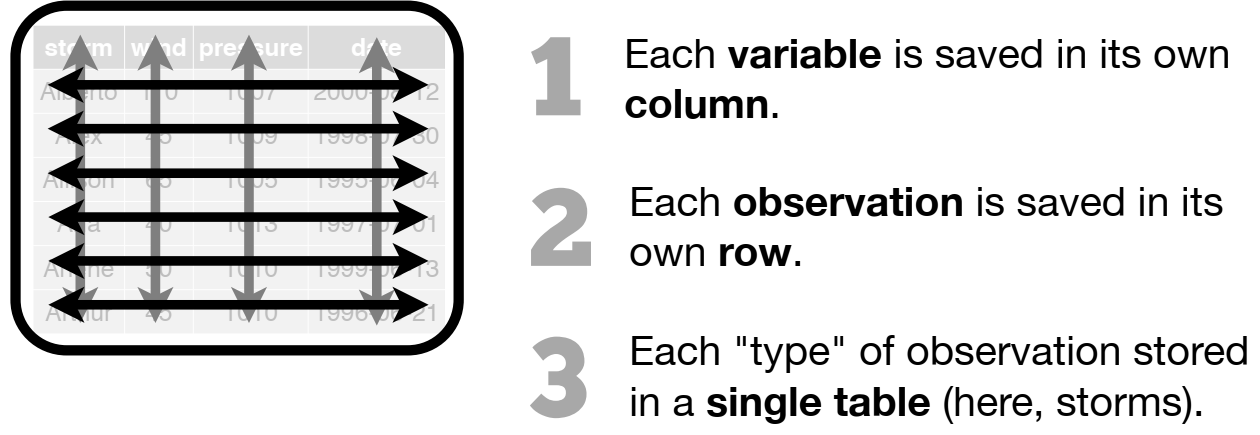
R functions like tidy data!
The keys to R’s superpowers are functions! There are four things you need to know to use a function:
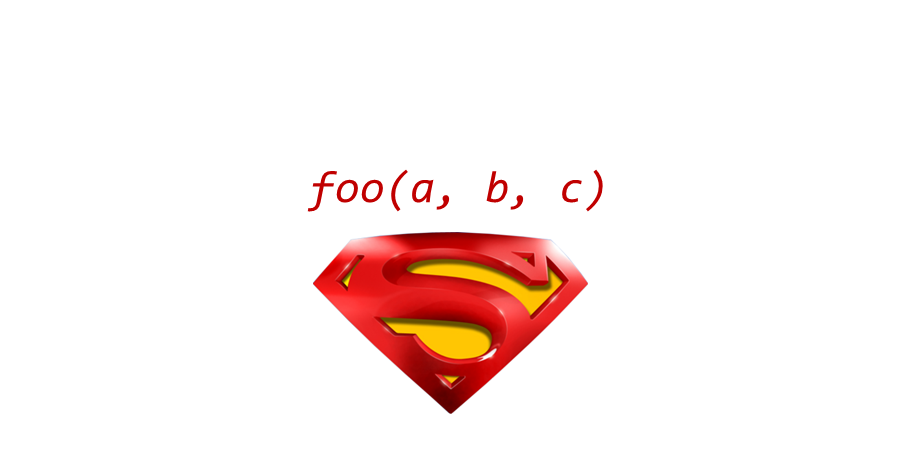



|
Finding the right R function, half the battle is. - Jedi MasteR Yoda |

|
Ask your friends
Ask Google / ChatGPT
Every function has a help page. To get to it enter ?
followed by the name of the function (without parentheses):
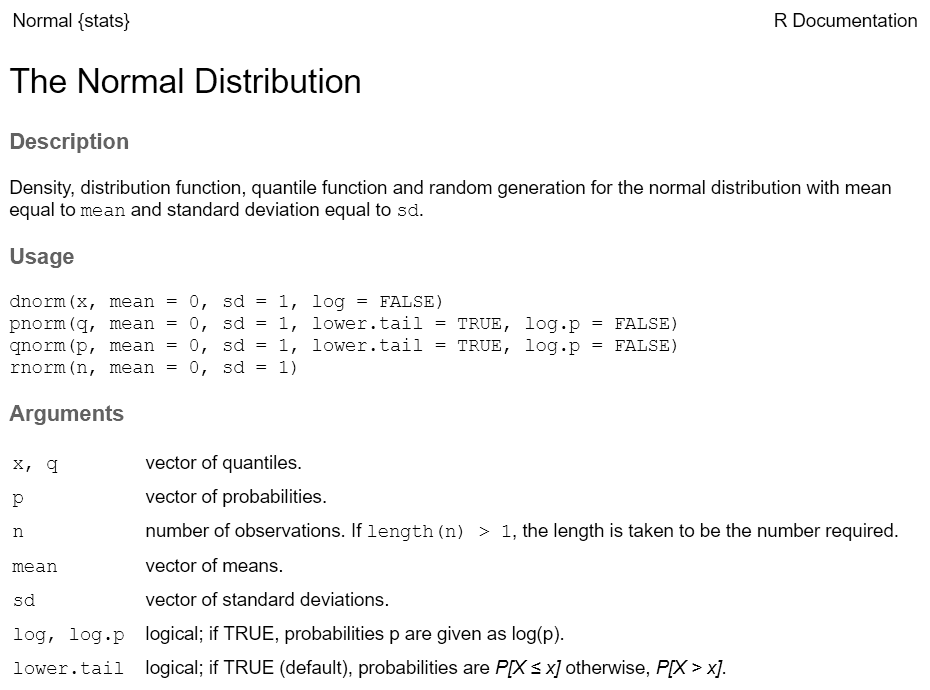
Most functions take arguments. Arguments can be required or optional (i.e. have a default value).
See the function’s help page to determine which arguments are expected. Example:
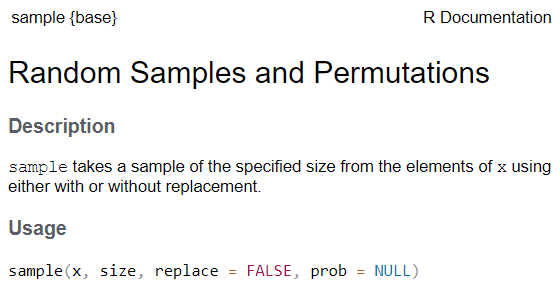
x and size have no default value → they are
required
replace and prob have default values → they
are optional
All arguments have names. You can explicitly name arguments when calling a function as follows:
Benefits of naming your arguments:
But you can omit argument names if you pass them in the order expected and don’t skip any.

Piping syntax is an alternative way of writing arguments into functions.
With piping, you use the pipe operator |> (or %>%) to ‘feed’ the result of one function into the next function.
Piping allows the results of one function to be passed as the first argument of the next function. Hence a series of commands to be written like a sentence.
Consider the expression:
zoo(moo(boo(foo(99)),n=4))


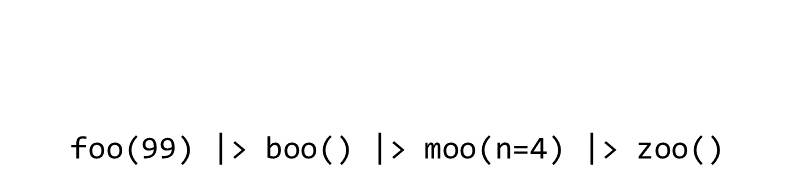
Keyboard shortcut for inserting the pipe operator:
ctrl + shift + m
You can tell RStudio which pipe to insert under Global Options >> Code
|> (‘native’ pipe
introduced R4.0)
%>% (from
magrittr package)
To split a chain of functions across multiple lines, end each line with a pipe operator:
If the receiving function requires additional arguments, just add them starting with the 2nd argument (or use named arguments):
Occasionally two or more packages will have different functions that
use the same name. 
When this happens, R will use whichever one was loaded first.
Best practice: use the package name and the :: reference
to specify which package a function is from.
When you use the package_name::function_name
syntax, you don’t actually have to first load the package with
library().
Resolving Name Conflicts with the conflicted
Package
When you call a function that exists in multiple packages, R uses whichever package was loaded first.
The conflicted package helps you avoid problems with
duplicate function names, by specifying which one to prioritize no
matter what order they were loaded.



These packages allow you to:
Example:
dplyr
An alternative (usually better) way to wrangle data frames than base R.
Part of the tidyverse.
Best way to familiarize yourself - explore the cheat sheet:
dplyr Functions| subset rows | filter(), slice() |
| order rows | arrange() |
| pick column(s) | select(), pull() |
| add new columns | mutate() |
Most dplyr functions take a tibble as the first
argument , and return a tibble.
This makes them very pipe friendly.
Look at the storms tibble:
## # A tibble: 6 × 13
## name year month day hour lat long status category wind pressure
## <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <fct> <dbl> <int> <int>
## 1 Amy 1975 6 27 0 27.5 -79 tropical de… NA 25 1013
## 2 Amy 1975 6 27 6 28.5 -79 tropical de… NA 25 1013
## 3 Amy 1975 6 27 12 29.5 -79 tropical de… NA 25 1013
## 4 Amy 1975 6 27 18 30.5 -79 tropical de… NA 25 1013
## 5 Amy 1975 6 28 0 31.5 -78.8 tropical de… NA 25 1012
## 6 Amy 1975 6 28 6 32.4 -78.7 tropical de… NA 25 1012
## # ℹ 2 more variables: tropicalstorm_force_diameter <int>,
## # hurricane_force_diameter <int>
Filter out only the records for category 3 or higher storms
storms |>
select(name, year, month, category) |> ## select the columns we need
filter(category >= 3) ## select just the rows we want## # A tibble: 1,262 × 4
## name year month category
## <chr> <dbl> <dbl> <dbl>
## 1 Caroline 1975 8 3
## 2 Caroline 1975 8 3
## 3 Eloise 1975 9 3
## 4 Eloise 1975 9 3
## 5 Gladys 1975 10 3
## 6 Gladys 1975 10 3
## 7 Gladys 1975 10 4
## 8 Gladys 1975 10 4
## 9 Gladys 1975 10 3
## 10 Belle 1976 8 3
## # ℹ 1,252 more rowsdplyr functions generally allow you to enter column
names without quotes.
In this exercise, we will:
Import some messy data from Excel
Wrangle the Penguins!
This exercise will be done in an R Notebook!
dplyr Pro Tipsselect()| select(-age) | Select all columns except age |
| select(fname:grade) |
Select all columns between fname and grade
|
| select(starts_with(“Sepal”)) |
Select all columns that start with ‘Sepal’ (see also ends_width(), contains(), and
matches()
|
my_dataframe |> rename(fname = First, lname = Last)
my_dataframe |> select(fname = First, lname = Last, term, grade, passed)
dplyr join functions| join data frames on a column | left_join(), right_join(), inner_join() |
| stack data frames | bind_rows() |
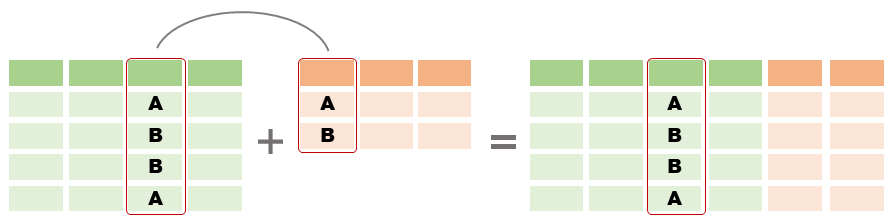
To join two data frames based on a common column, you can use:
left_join(x, y, by)
where x and y are data frames, and by is the name of a column they have in common.
If there is only one column in common, and if it has the same name in both data frames, you can omit the by argument.
If the common column is named differently in the two data frames, you can deal with that by passing a named vector as the by argument. See below.
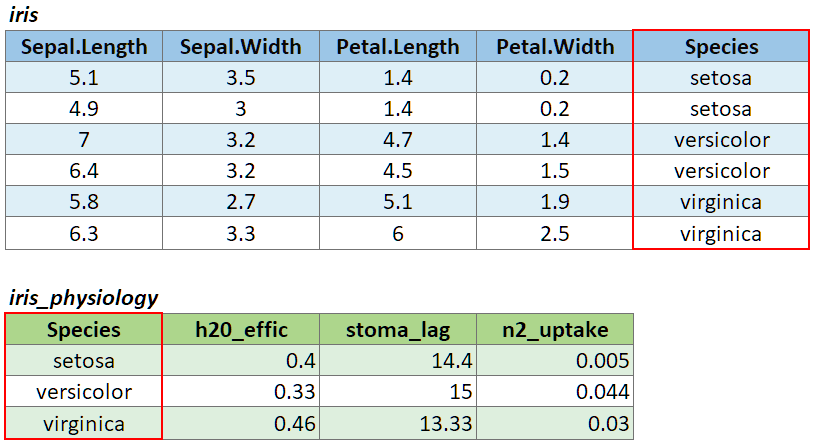
To illustrate a table join, we’ll first import a csv with some fake data about the genetics of different iris species:
# Create a data frame with additional info about the three IRIS species
iris_genetics <- data.frame(Species=c("setosa", "versicolor", "virginica"),
num_genes = c(42000, 41000, 43000),
prp_alles_recessive = c(0.8, 0.76, 0.65))
iris_genetics## Species num_genes prp_alles_recessive
## 1 setosa 42000 0.80
## 2 versicolor 41000 0.76
## 3 virginica 43000 0.65We can join these additional columns to the iris data frame with
left_join():
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species num_genes
## 1 5.1 3.5 1.4 0.2 setosa 42000
## 2 4.9 3.0 1.4 0.2 setosa 42000
## 3 4.7 3.2 1.3 0.2 setosa 42000
## 4 4.6 3.1 1.5 0.2 setosa 42000
## 5 5.0 3.6 1.4 0.2 setosa 42000
## 6 5.4 3.9 1.7 0.4 setosa 42000
## 7 4.6 3.4 1.4 0.3 setosa 42000
## 8 5.0 3.4 1.5 0.2 setosa 42000
## 9 4.4 2.9 1.4 0.2 setosa 42000
## 10 4.9 3.1 1.5 0.1 setosa 42000
## prp_alles_recessive
## 1 0.8
## 2 0.8
## 3 0.8
## 4 0.8
## 5 0.8
## 6 0.8
## 7 0.8
## 8 0.8
## 9 0.8
## 10 0.8
If you need to join tables on multiple columns, add additional column
names to the by argument.
Join columns must be the same data type (i.e., both numeric or both character).
There are several variants of left_join(), the most
common being right_join() and
inner_join(). See help for details.
If the join column is named differently in the two tables, you can
pass a named character vector as the by argument. A named
vector is a vector whose elements have been assigned names. You can
construct a named vector with c().
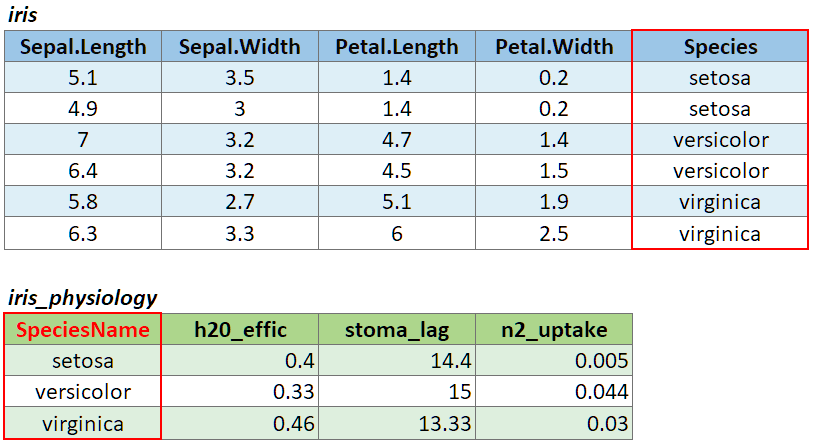
For example if the join column was named ‘SpeciesName’ in x, and just ‘Species’ in y, your expression would be:
left_join(x, y, by = c("SpeciesName" = "Species"))
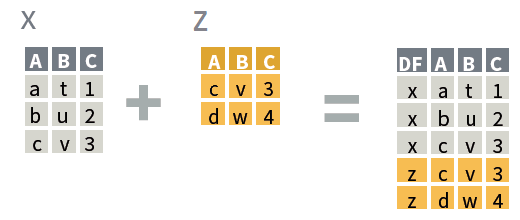
bind_rows(x, y)
where x and
y:

Reshaping data includes:
The go-to Tidyverse package for reshaping data frames is tidyr
|
Step 1 (optional): group rows (i.e., change the unit of analysis) |
group_by() |
|
Step 2: Compute summaries for each group of rows |
Add summary columns with summarize() Aggregate functions available: n(), mean(), median(), sum(), sd(), IQR(), first(), etc. |
Example:
## # A tibble: 6 × 13
## name year month day hour lat long status category wind pressure
## <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <fct> <dbl> <int> <int>
## 1 Amy 1975 6 27 0 27.5 -79 tropical de… NA 25 1013
## 2 Amy 1975 6 27 6 28.5 -79 tropical de… NA 25 1013
## 3 Amy 1975 6 27 12 29.5 -79 tropical de… NA 25 1013
## 4 Amy 1975 6 27 18 30.5 -79 tropical de… NA 25 1013
## 5 Amy 1975 6 28 0 31.5 -78.8 tropical de… NA 25 1012
## 6 Amy 1975 6 28 6 32.4 -78.7 tropical de… NA 25 1012
## # ℹ 2 more variables: tropicalstorm_force_diameter <int>,
## # hurricane_force_diameter <int>
For each month, how many storm observations are saved in the data frame
storms |>
select(name, year, month, category) |> ## select the columns we need
group_by(month) |> ## group the rows by month
summarize(num_storms = n()) ## for each group, report the count## # A tibble: 10 × 2
## month num_storms
## <dbl> <int>
## 1 1 70
## 2 4 66
## 3 5 201
## 4 6 809
## 5 7 1651
## 6 8 4442
## 7 9 7778
## 8 10 3138
## 9 11 1170
## 10 12 212