
Andy
Lyons
October 4, 2023

R as a fancy calculator:
(pi
* 5 ^ 2) / sqrt(4 / 3)
Saving results as objects:
my_volume = (4 / 3) * pi * (1.5 ^ 3)
All R objects have a class (e.g., numeric, character, data frame)
Objects that contain multiple values of the same class are called vectors
Some functions that return
vectors:
seq()
rnorm()
sample()
You can build vectors from scratch using:
c()
You can subset or extract elements from vectors using square
brackets:
With a
vector of integers (indices):
LETTERS[1:4]
Logical vector:
LETTERS[
c(T,T,T,T,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F,F)
]
Logical expression:
x =
1:100
x [x %% 3 == 0]
Functions and operators that do something to each element of a
vector and return another vector are said to be
vectorized
+ - * /
round(), abs()
Functions that take multiple elements of a vector and spit out a
single value are said to be aggregate
functions
sum(), min(), mean(), max()
R comes with some basic plot functions:
hist(x)
boxplot(x)
plot(x, y) ## scatterplot
plot(x, y, type = 'b') ## 'b' = plot both points and lines
First half
Second half

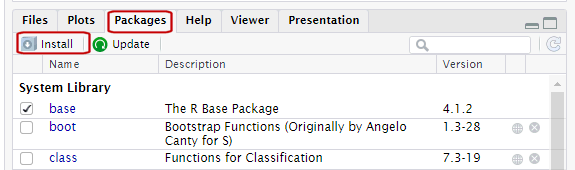
Packages are what R calls extensions or add-ons.
Three simple steps to use the functions in a package:
Figure out which package you need
Install (i.e., download) it (just once)
The keys to R’s superpowers are functions! There are four things you need to know to use a function:
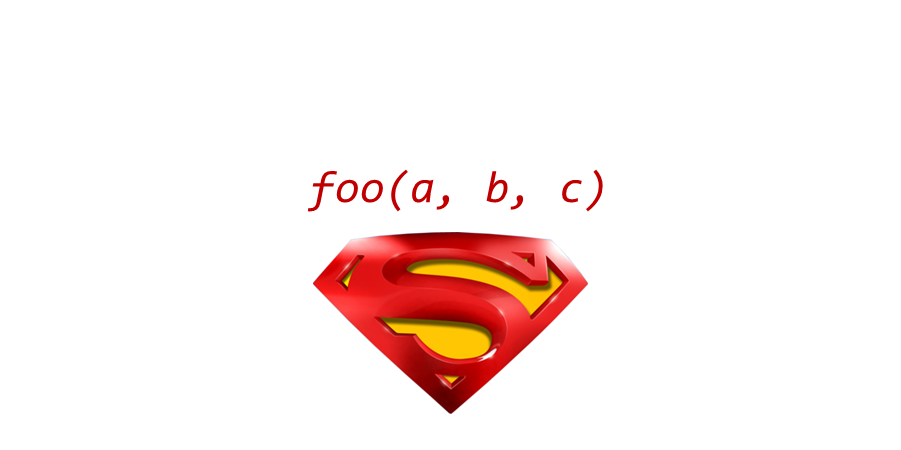



|
Finding the right R function, half the battle is. - Jedi MasteR Yoda |

|
Ask your friends
Ask Google
Cheatsheets!
Every function has a help page. To get to it enter ?
followed by the name of the function (without parentheses):
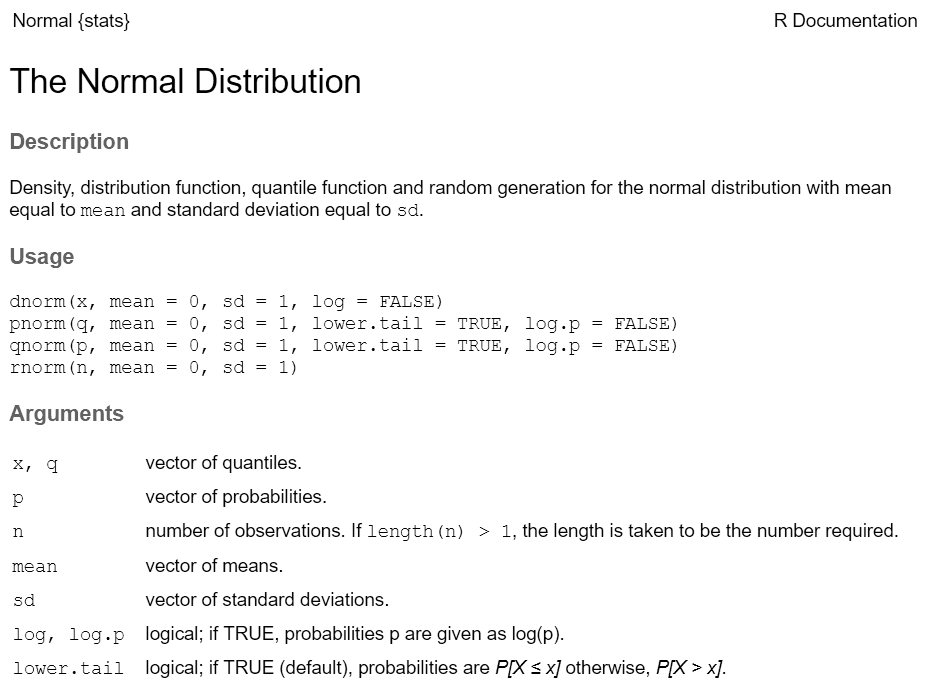
Most functions take arguments. Arguments can be required or optional (i.e. have a default value).
See the function’s help page to determine which arguments are expected. Example:
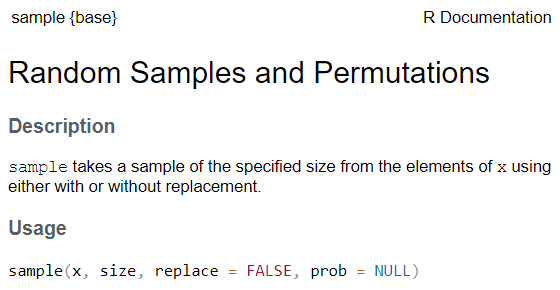
x and size have no default value → they are
required
replace and prob have default values → they
are optional
All arguments have names. You can explicitly name arguments when calling a function as follows:
Benefits of naming your arguments:
But you can omit argument names if you pass them in the order expected and don’t skip any.

Piping syntax is an alternative way of writing arguments into functions.
With piping, you use the pipe operator |> (or %>%) to ‘feed’ the result of one function into the next function.
Piping allows the results of one function to be passed as the first argument of the next function. Hence a series of commands to be written like a sentence.
Consider the expression:
zoo(moo(boo(foo(99)),n=4))


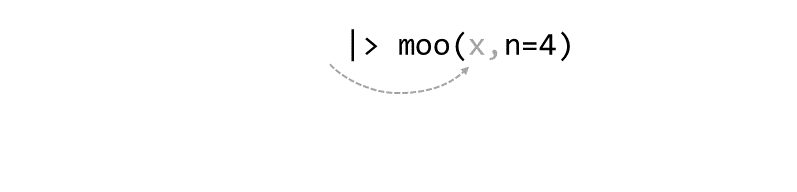

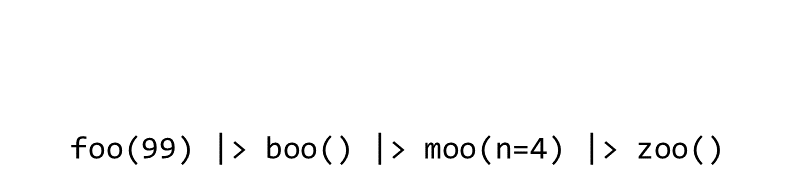
Keyboard shortcut for inserting the pipe operator:
ctrl + shift + m
You can tell RStudio which pipe to insert under Global Options >> Code
|> (‘native’ pipe
introduced R4.0)
%>% (from
magrittr package)
To split a chain of functions across multiple lines, end each line with a pipe operator:
If the receiving function requires additional arguments, just add them starting with the 2nd argument (or use named arguments):
Exercise 3 Topics
library()
R has two data classes that organize data in rows and columns:
rows
aka:
- record
- case
- feature (spatial)
columns
aka: variable, field
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
To get an individual column, use $
## Girth Height Volume
## 1 8.3 70 10.3
## 2 8.6 65 10.3
## 3 8.8 63 10.2
## 4 10.5 72 16.4
## 5 10.7 81 18.8
## 6 10.8 83 19.7## [1] 70 65 63 72 81 83 66 75 80 75 79 76 76 69 75 74 85 86 71 64 78 80 74 72 77
## [26] 81 82 80 80 80 87## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 63 72 76 76 80 87
You can use square bracket notation or tidyverse methods (dplyr).
Come back for Part 3. Data Wrangling
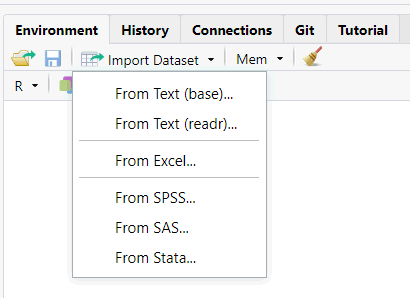
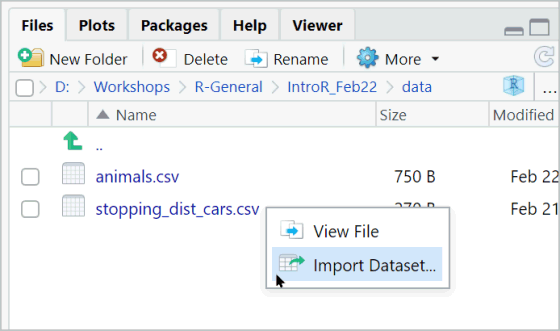
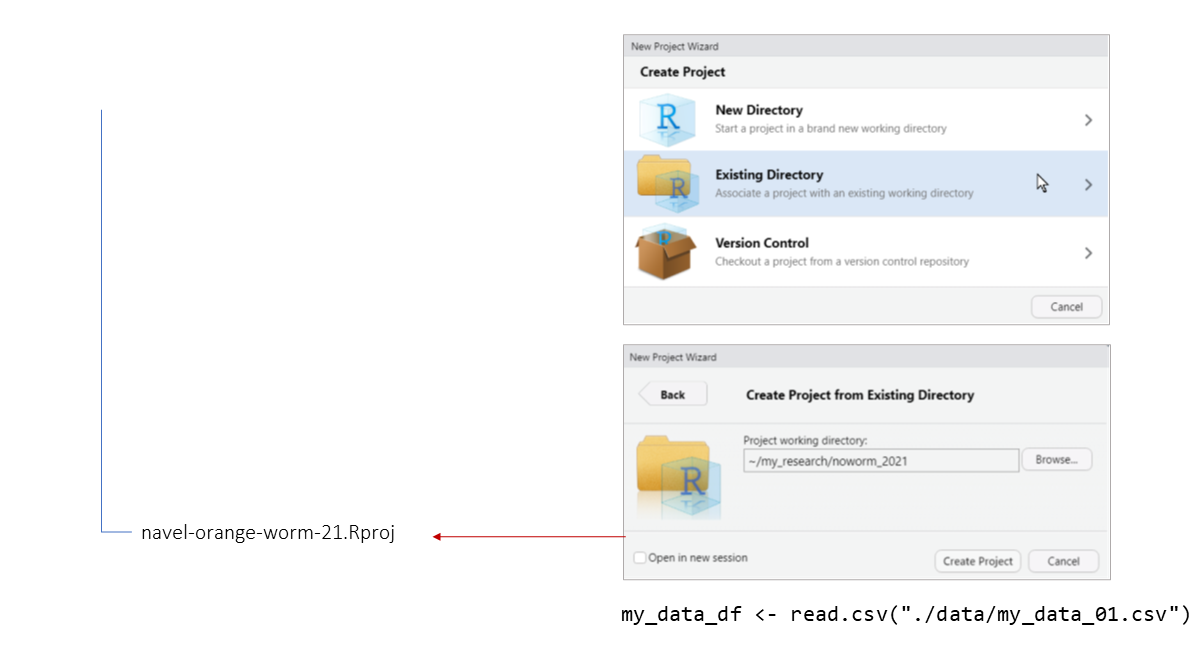
When you import (or export) data with code, you have to provide a path to a file.
Paths can be absolute (full), or relative working directory.
Complete path:
Windows users: Beware the slashes!
The following will not work beause of the slashes:
Back slashes must be converted to one of the following:
C:/noworm/trials/data
C:\\noworm\\trials\\data
Even though you don’t see it, there is always working directory.
You can view the current working directory with:
Changing the working directory is easy!
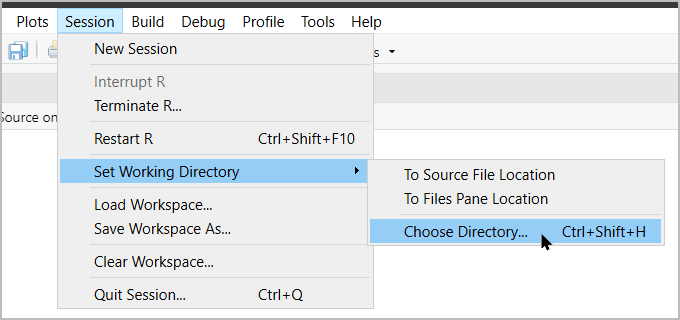
…or with code:
When using relative paths, the first '.' represents the
working directory.
The main advantage of using relative paths is portability of your project.




Exercise 4 Topics