
Andy
Lyons
October 18, 2023


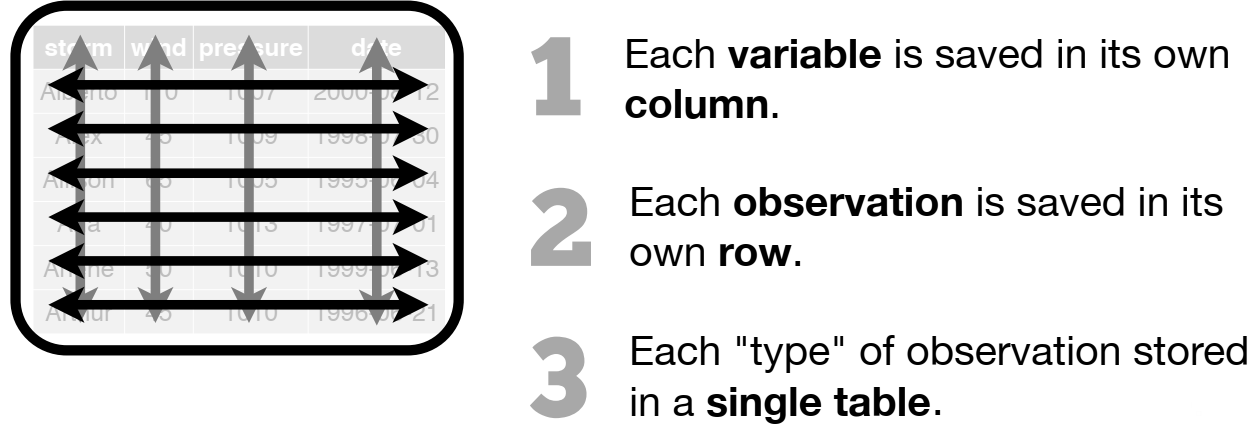



dplyr)| subset rows | filter(), slice() |
| order rows | arrange() |
| pick column(s) | select(), pull() |
| add new columns | mutate() |
dplyr)| join data frames on a column | left_join(), right_join(), inner_join() |
| stack data frames | bind_rows() |
tidyr)| convert rows to columns | pivot_wider() |
| turn columns into rows | pivot_longer() |
dplyr)|
Step 1 (optional): group rows (i.e., change the unit of analysis) |
group_by() |
|
Step 2: Compute summaries for each group of rows |
summarize() with: n(), mean(), median(), sum(), sd(), first(), etc. |
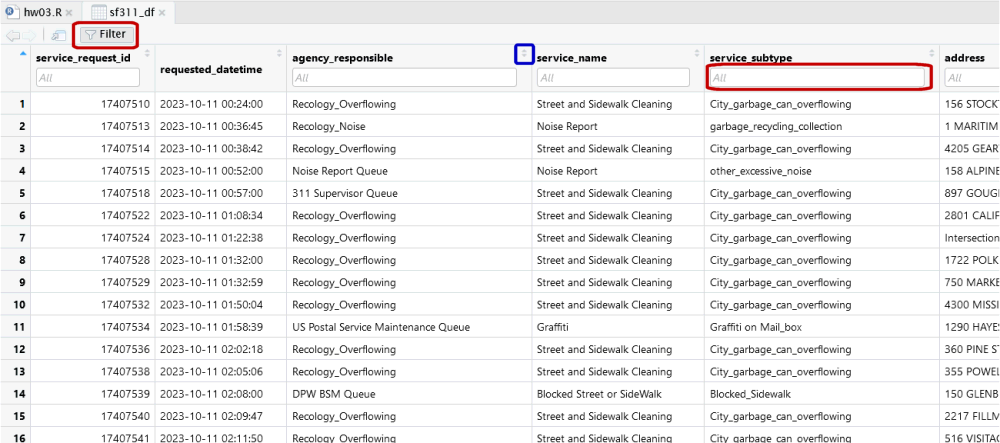
Exploring large data frames with the View pane:
Date class in R: Date
Current date:
## [1] "Date"## [1] "2023-10-29"
Text to date:
## [1] "2023-09-25"
Date & time classes: POSIXct,
POSIXlt
Functions from lubridate:
ymd_hms(), ymd_hm(), ymd_h()
ydm_hms(), ydm_hm(), ydm_h()
mdy_hms(), mdy_hm(), mdy_h()
dmy_hms(), dmy_hm(), dmy_h()
Examples:
## [1] "2017-11-28 14:02:00 UTC"## [1] "2017-11-28 14:02:00 PST"
Pro Tips:
To see the accepted time zone names, run
OlsonNames()
Don’t name an object ‘date’
## [1] "2023-10-18"## [1] "Date"
## [1] "2023-10-18 17:15:00 UTC"## [1] "POSIXct" "POSIXt"
summary()
is.na(), !is.na()
tidyr::drop_na()
common replacement choices: 0, mean, median, etc.
mutate( new_col = if_else(…) )
tidyr::replace_na()
mutate( new_col = if_else(…) )
mutate( new_col = coalesce(…) )
Sample methods:

https://posit.cloud/content/6638058

The default time zone on Posit Cloud is UTC.
To mimic your local computer, you can change the default time zone:
To create a new column which includes lagged values, you can use
lag()
lead()
Best Option: Use a file format that supports date and time classes
Examples:
Second best: Use unambiguous character formatting
For file formats that don’t enforce data types (e.g., csv, txt, Excel), save dates as text formatted as:
yyyy-mm-dd
Times can be formatted as ISO 8601. Example:
"2023-10-29T14:30:11-0700"
Note: the last five characters encode the time zone:
-0700.
lubridate makes this easy with
format_ISO8601()
## [1] "2023-11-10T10:09:01-0800"
Load Palmer Penguins data frame:
## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <fct> <fct> <dbl> <dbl> <int> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## # ℹ 2 more variables: sex <fct>, year <int>
Use ggplot to make a scatter plot:







aes()
ggplot(penguins, aes(x = flipper_length_mm , y = bill_length_mm , color = species)) +
geom_point() +
ggtitle("Bill Length vs Flipper Length for 3 Species of Penguins")
aes() sets the default source for each visual property
(or aesthetic) of the plot layers
x - where it falls along the x-axis
y - where it falls along the y-axis
color
fill
size
geom_xxxx() functions you useaes() the visual properties you want linked
to the datageom_xxxx() functions add layers
drawn from the bottom up
some common geoms:
geom_point(col = pop_size)
geom_point(col = “red”)
visual properties are inherited (from
aes())
each geom has default color palettes and legend settings
In the example below, note where geom_boxplot() gets its
visual properties:
aes()ggplot(penguins, aes(x = species, y = bill_length_mm)) +
geom_boxplot(color = "navy", fill = "yellow", size = 1.5)## Warning: Removed 2 rows containing non-finite values (`stat_boxplot()`).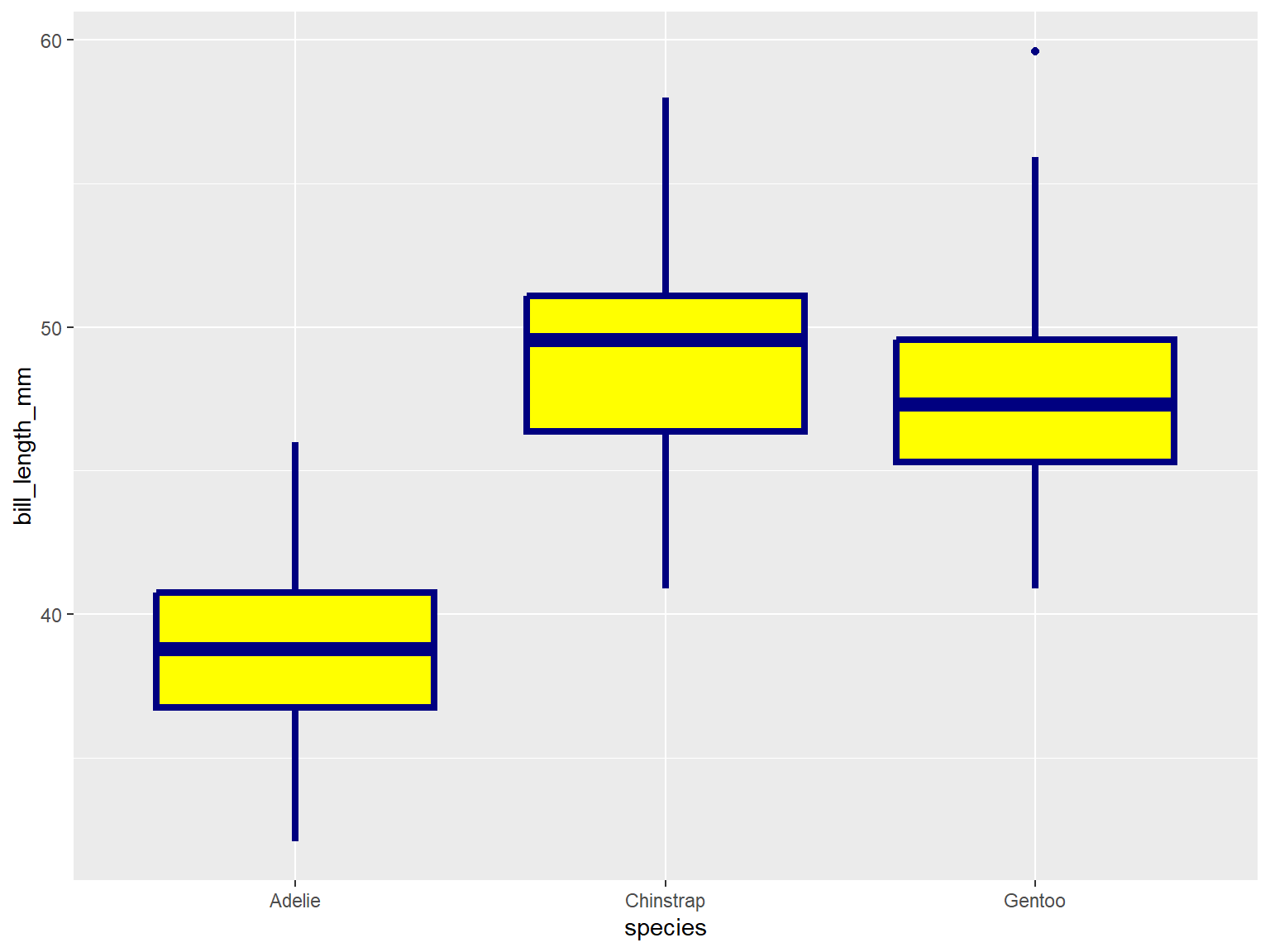
geom_xxxx() functions can also be used to add other
graphic elements:
ggplot(penguins, aes(x = species, y = bill_length_mm)) +
geom_boxplot(color = "navy", fill = "yellow", size = 1.5) +
geom_hline(yintercept = 43.9, size=3, color="red") +
geom_label(x = 3, y = 58, label = "Gentoo penguins \n are just the best!")## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.## Warning: Removed 2 rows containing non-finite values (`stat_boxplot()`).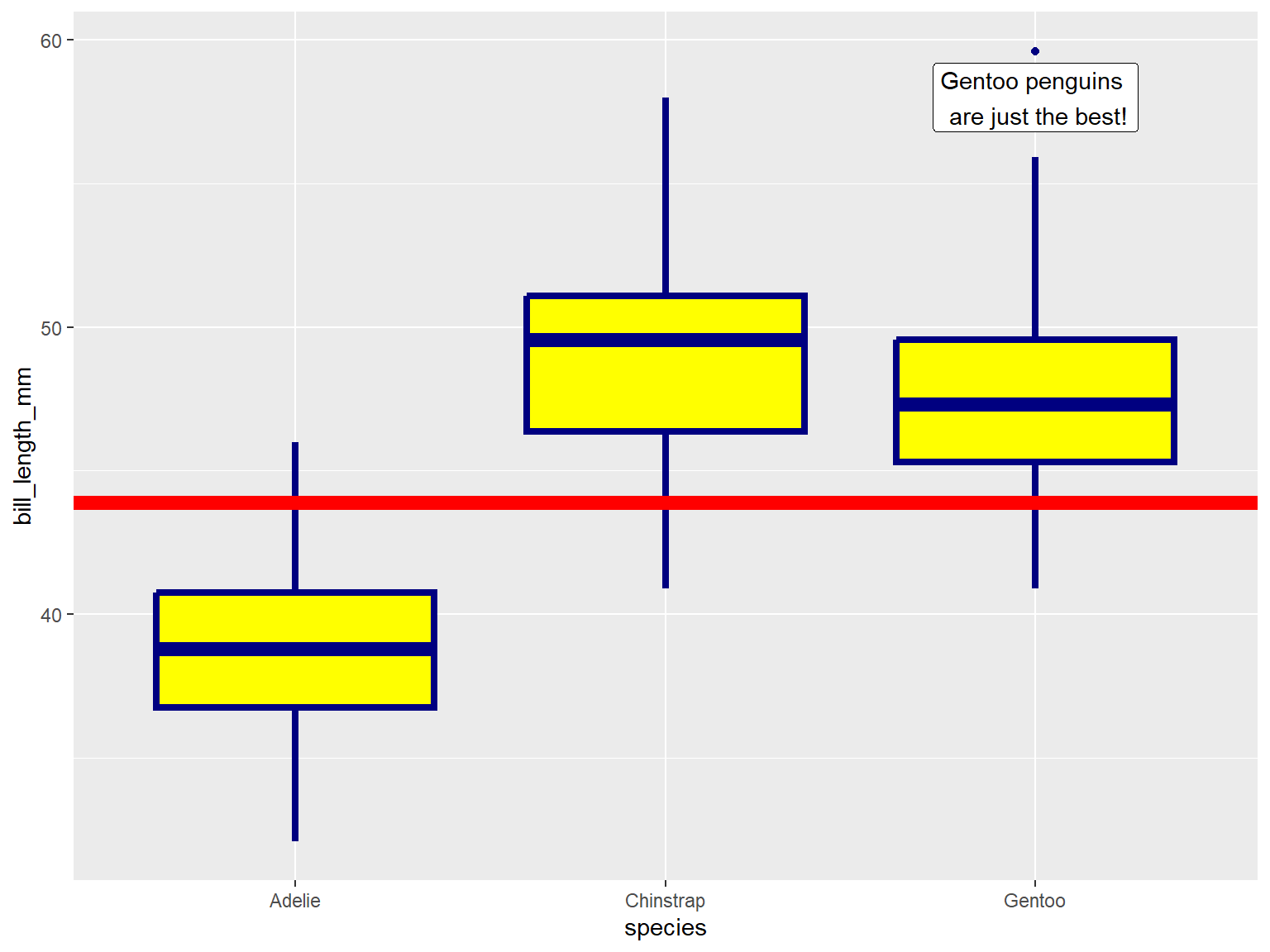
Done!