December
2,
2022
https://ucanr-igis.github.io/agroclimR/





Type of Organization
Email domain
Location
Familiarity with Agroclimate Metrics
Familiarity with R
Develop general familiarity with:
Come away with:
Workshop materials:
Agroclimate metrics reflect weather driven factors that influence the development of plants and insects.
Examples:
These are abiotic factors but they influence biotic factors that affect crops (like disease).
The main ingredients of metrics are measurements of weather variables (like air temperature).
We can compute them for the past, present, and future.
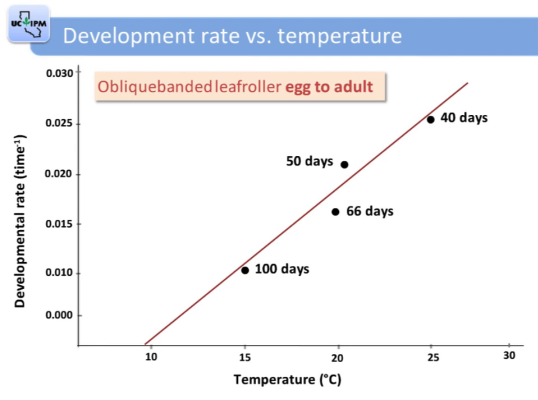
Pistachio nut embryo length vs
degree days
Zhang et al (2021)

 https://doi.org/10.1016/j.ufug.2018.07.020 |
Where can I go today to see the climate my city will have 50 years from now? |
| How many generations of Navel Orangeworm will growers have to deal with in the coming decades? |
 https://doi.org/10.1016/j.scitotenv.2020.142657 |
 https://doi.org/doi:10.1371/journal.pone.0020155 |
How much chill can we expect in the coming decades? Will there be enough to have a economically viable farming operation? |
| What kind of frost exposure will we see mid-century? |
 https://doi.org/10.1016/j.scitotenv.2020.143971 |
 https://doi.org/10.3390/agronomy12010205 |
What is the ‘new normal’ for agroclimatic metrics for specialty crops? |
R is an extremely flexible computing environment,
with
strengths in:





owmr:
OpenWeatherMap API Wrapper
rnoaa: ‘NOAA’
Weather Data from R
riem: Accesses
Weather Data from the Iowa Environment Mesonet
…plus many
others
Or write your own!

File server
API service

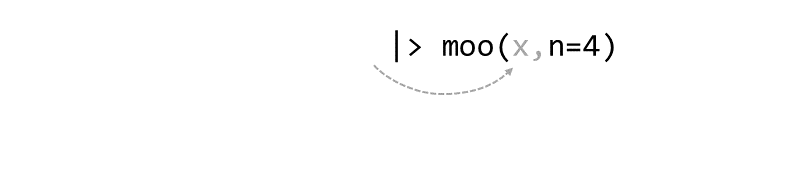
Piping syntax is an alternative way of chaining functions together than nested parentheses:
zoo(moo(boo(foo(99),k=7),n=4))
With piping, you use the pipe operator |> or %>% to ‘feed’ the result of one function as the first argument to the next function.

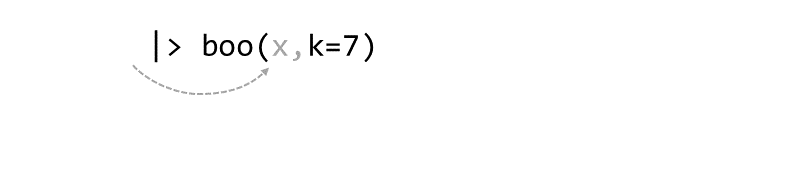

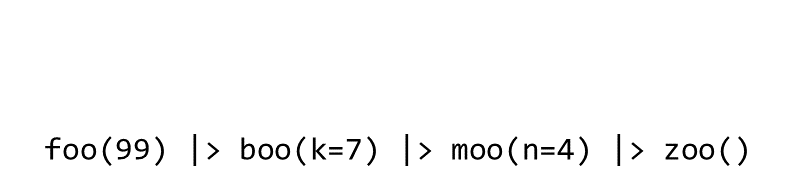
ctrl + shift + m|> (‘native’
pipe introduced R4.0)%>% (traditional, requires
magrittr package)
Whatever is needed to get your data frame ready for the
function(s)
you need for analysis and visualization.

The go-to package for wrangling data frames is dplyr:
dplyr Functions| subset rows | filter(), slice() |
| order rows | arrange(), arrange(desc()) |
| pick column(s) | select(), pull() |
| add new columns | mutate() |
| offset rows | lead(), lag() |
| vectorized conditional checks | if_else(), case_when() |
| join data frames | left_join(), right_join(), inner_join() |
Most dplyr functions take a tibble, and
return a tibble.
This makes them very pipe friendly.
Let’s look at the storms tibble (NOAA Atlantic hurricane database, 1975-2000, including position, wind speed, etc. every six hours):
## # A tibble: 6 × 13
## name year month day hour lat long status categ…¹ wind press…² tropi…³
## <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr> <ord> <int> <int> <int>
## 1 Amy 1975 6 27 0 27.5 -79 tropi… -1 25 1013 NA
## 2 Amy 1975 6 27 6 28.5 -79 tropi… -1 25 1013 NA
## 3 Amy 1975 6 27 12 29.5 -79 tropi… -1 25 1013 NA
## 4 Amy 1975 6 27 18 30.5 -79 tropi… -1 25 1013 NA
## 5 Amy 1975 6 28 0 31.5 -78.8 tropi… -1 25 1012 NA
## 6 Amy 1975 6 28 6 32.4 -78.7 tropi… -1 25 1012 NA
## # … with 1 more variable: hurricane_force_diameter <int>, and abbreviated
## # variable names ¹category, ²pressure, ³tropicalstorm_force_diameter
Which category 3 hurricane had the largest diameter of hurricane force winds?
We can answer this question in one easy-to-read expression:
storms %>%
select(name, year, month, day, hour, category, status, hurricane_force_diameter) |> ## select the columns we need
filter(category == 3) |> ## category 3
arrange(desc(hurricane_force_diameter)) |> ## arrange descending by diameter
slice(1:3) ## show the top 3## # A tibble: 3 × 8
## name year month day hour category status hurricane_force_diameter
## <chr> <dbl> <dbl> <int> <dbl> <ord> <chr> <int>
## 1 Ivan 2004 9 16 0 3 hurricane 165
## 2 Ivan 2004 9 16 6 3 hurricane 165
## 3 Nicole 2016 10 13 15 3 hurricane 160Note also that with dplyr functions you generally don’t have to put column names in quotes.
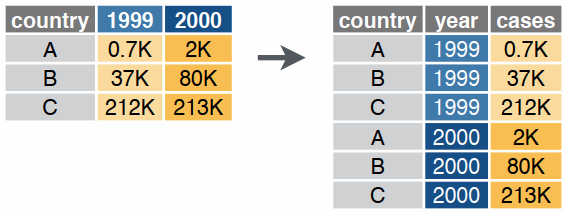
Reshaping data includes:
The go-to tidyverse package for reshaping data frames is tidyr:
tidyr Functionspivot_wider()
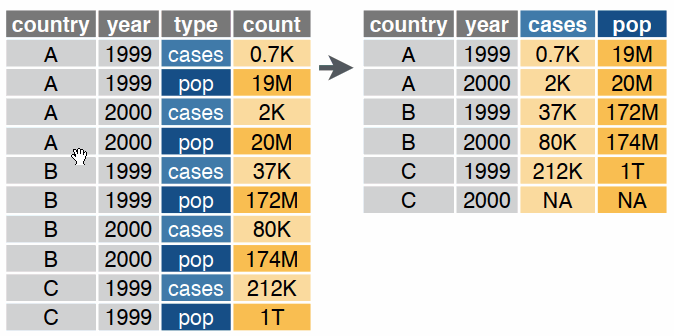
pivot_longer()
Agroclimate metrics frequently requires working with date values.
These are not Date objects:
To convert date-formatted text to Date objects use
as.Date():
For everything else…
rle()Heat spells or heat waves are often defined as number of consecutive days where the maximum temperature exceeds a certain threshold.
It is relatively easy to flag which days exceed a threshold:
We can pull out consecutive days of high temperatures with the
rle() function.
rle()Run length encoding is a way to compress data that is i) ordered, and ii) has repetitions. The best way to see how it works is with an illustration:
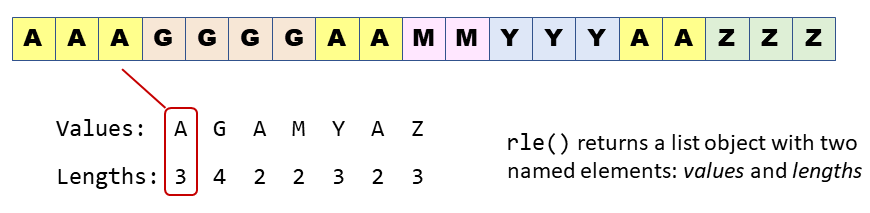
Now imagine we have time series data where TRUE means a hot day:
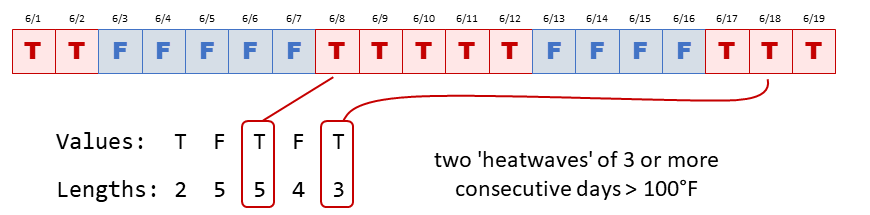
To compute this with rle():
hotdays_df <- data.frame(dt = seq(from = as.Date("2020-06-01"), to = as.Date("2020-06-19"), by = 1),
hotyn = c(T,T,F,F,F,F,F,T,T,T,T,T,F,F,F,F,T,T,T))
hotdays_df## dt hotyn
## 1 2020-06-01 TRUE
## 2 2020-06-02 TRUE
## 3 2020-06-03 FALSE
## 4 2020-06-04 FALSE
## 5 2020-06-05 FALSE
## 6 2020-06-06 FALSE
## 7 2020-06-07 FALSE
## 8 2020-06-08 TRUE
## 9 2020-06-09 TRUE
## 10 2020-06-10 TRUE
## 11 2020-06-11 TRUE
## 12 2020-06-12 TRUE
## 13 2020-06-13 FALSE
## 14 2020-06-14 FALSE
## 15 2020-06-15 FALSE
## 16 2020-06-16 FALSE
## 17 2020-06-17 TRUE
## 18 2020-06-18 TRUE
## 19 2020-06-19 TRUE
First, run the vector in rle():
Now we can write R expressions to pull out what we want.
## Which groups meet both conditions to be a heat wave?
group_is_heatwave <- (x$values == TRUE) & (x$lengths >= 3)
group_is_heatwave## [1] FALSE FALSE TRUE FALSE TRUE## [1] 2
## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_l…¹ body_…² sex year
## <fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
## 2 Adelie Torgersen 39.5 17.4 186 3800 fema… 2007
## 3 Adelie Torgersen 40.3 18 195 3250 fema… 2007
## 4 Adelie Torgersen NA NA NA NA <NA> 2007
## 5 Adelie Torgersen 36.7 19.3 193 3450 fema… 2007
## 6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
## # … with abbreviated variable names ¹flipper_length_mm, ²body_mass_g
Make a scatter plot:
ggplot(penguins, aes(x = flipper_length_mm, y = bill_length_mm, color = species)) +
geom_point() +
ggtitle("Bill Length vs Flipper Length for 3 Species of Penguins")## Warning: Removed 2 rows containing missing values (`geom_point()`).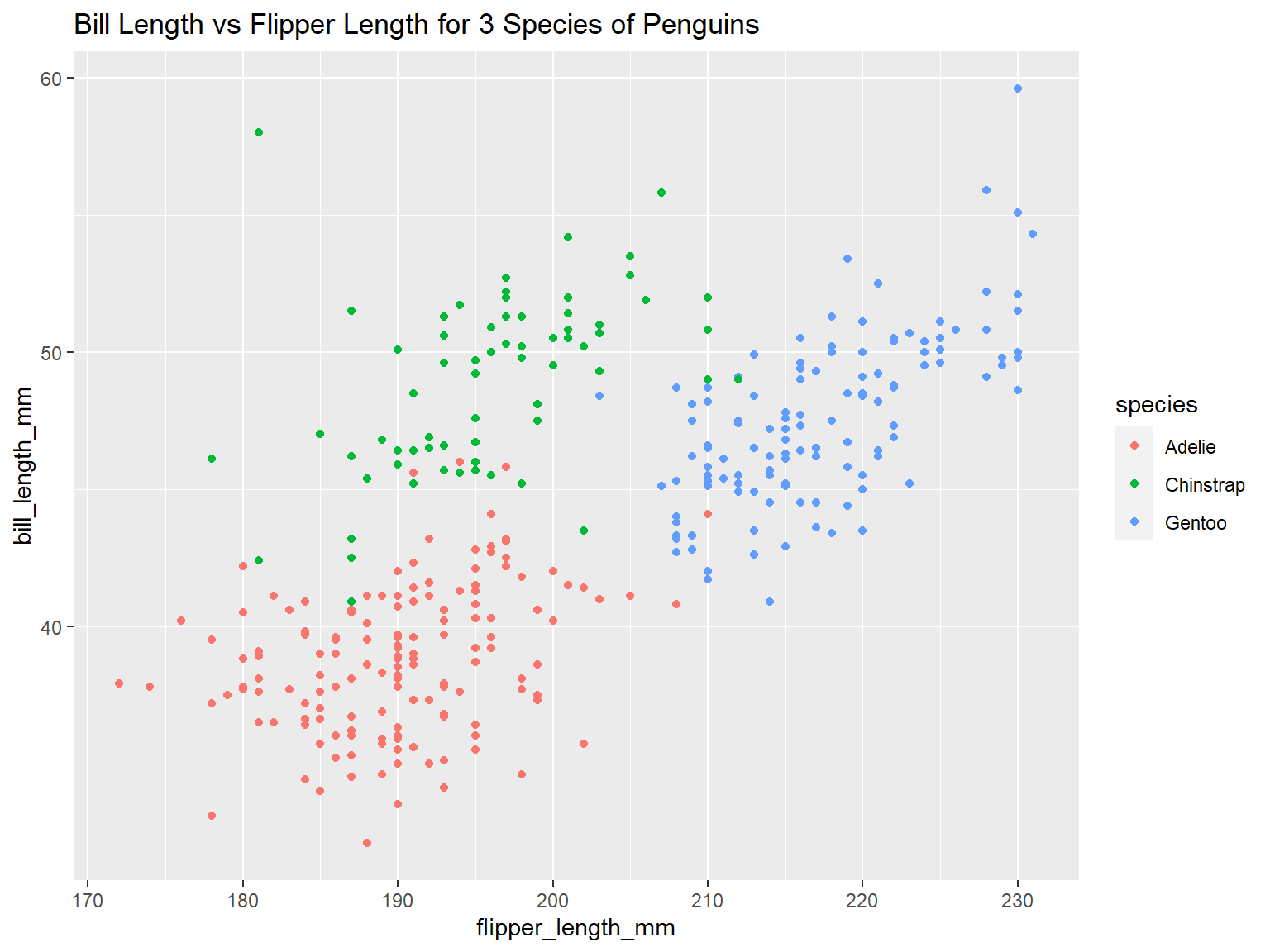







R Notebooks are written in “R Markdown”, which combines text and R code.
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
RStudio Cloud project for this workshop:
https://posit.cloud/content/5055980
After it opens:

Or see the Completed Notebook #1.